Research: Software
The mHealth group is committed to releasing software as often as possible. Generally, we want to make as much of our code available as possible, especially for published algorithms (see the Datasets page).
We currently have two open-source applications that may be of particular interest to the research community:
We currently have two open-source applications that may be of particular interest to the research community:
Signaligner Pro: Supporting Multi-day Raw Sensor Data Annotation
|
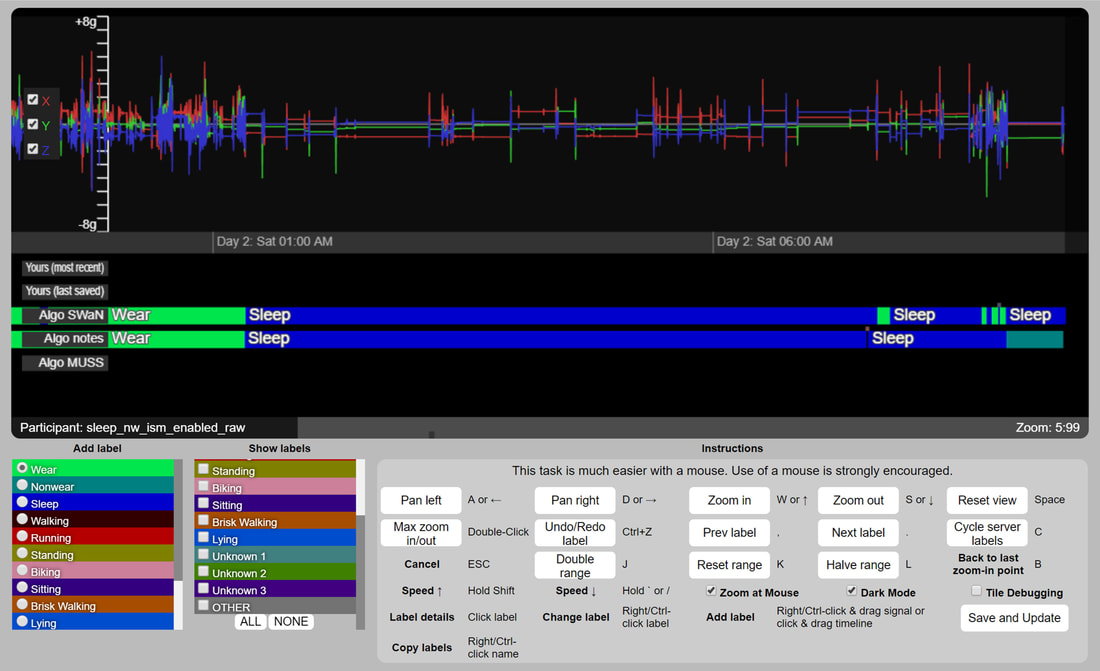
We have repurposed our prior work on human computation games to crowdsource raw sensor data annotations into an interactive tool for experts. We developed this tool to enable activity recognition, physical activity, and sleep behavior researchers interested in using raw sensor data to explore and annotate high-sampling rate raw data collected on individuals for a week of data or more. We have found such a tool to be vital in our own work, and so we have made it available to others and continue to improve it. Currently, the tool is being used to label data for prolonged physical activities and everyday activities, including sleep, but also short lived activities such as hand washing and face touching.
The tool and documentation is available on GitHub: https://crowdgames.github.io/signaligner/ Collaborators: Seth Cooper (Northeastern) |
MIX{Wild}: Supporting Mixed Model Analysis with Longitudinal Data
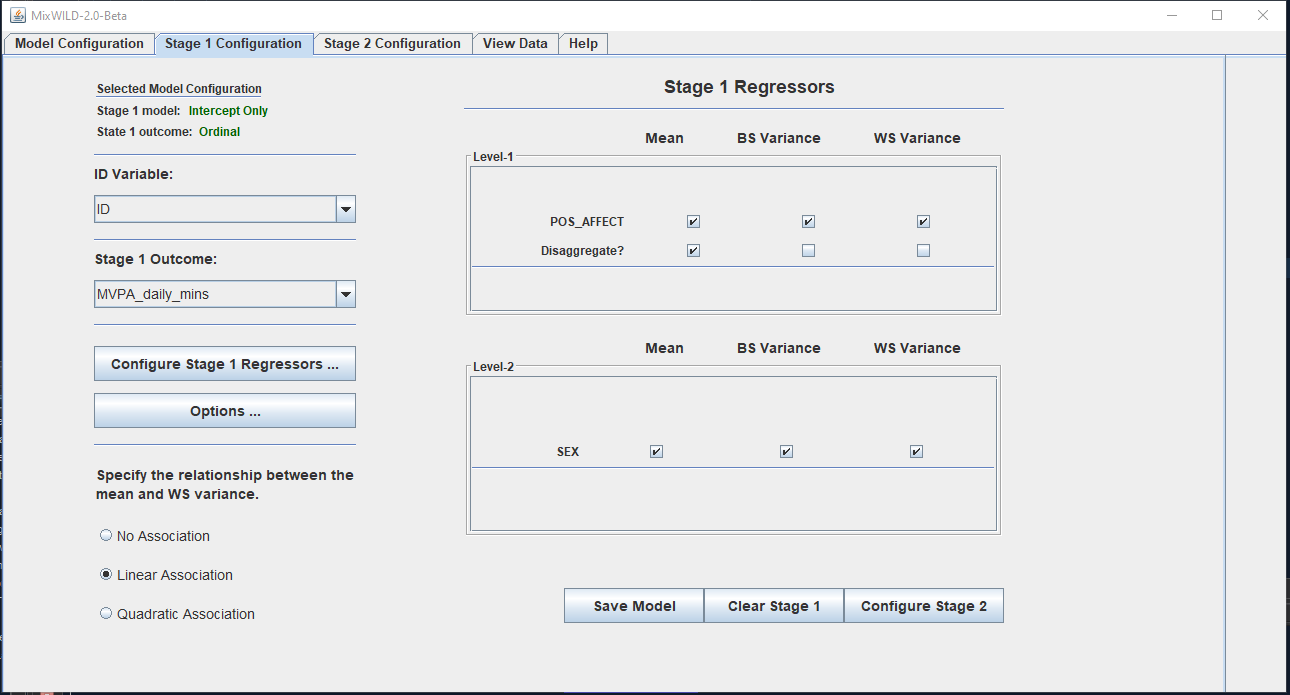
We are developing a free and open-source software application called MixWILD (“Mixed Effects Modeling With Intensive Longitudinal Data”). The software is for examining the effects of variance and slope of time-varying variables in intensive longitudinal data, especially from data collected using ecological momentary assessment. It is designed with user-friendly desktop graphical user interface (GUI) that allows researchers to build complex multilevel models without programming. The software is being developed with NIH/NCI funding.
The tool and documentation is available on GitHub: https://reach-lab.github.io/MixWildGUI/
The tool and documentation is available on GitHub: https://reach-lab.github.io/MixWildGUI/
Collaborators: University of Chicago, University of Southern California
Supported by: NIH/NCI